It's 6:30 on a Saturday morning and I'm up earlier than I usually am on a work day for some reason.
I have been slaving away at learning the extremely detailed language of a phylogenetic analysis software platform called RevBayes. The kinds of analysis this software is capable of doing are very sophisticated and useful, but the Byzantine labyrinth of calls, errors, bugs, troubleshooting, and repeated test runs has been exhausting for about two weeks now. On top of that, the resources used by the analyses are extraordinary. I bought a new laptop designed for gaming, which has 16GB RAM and 8 cores, just to be able to run test files. In addition to the computer upgrade, and at the same time that I have been learning RevBayes, I've been learning how to interact with the Agave computing cluster at ASU, which in itself has been a very steep learning curve. But I finally worked it all out to prepare a working script in RevBayes and submit a job to the scheduler on Agave. The scheduler is called "Slurm," and this has become a running joke between M and me, regarding anything gross, or the evil villain Dr. Slurm, etc. Submitting a job successfully has involved a ton of trial and error learning two huge new languages, sbatch and srun. Of course, RevBayes uses a more complex, multithreading platform called mpi, and submitting an mpi job involves a ton of extra strange complications, etc.
Meanwhile, RevBayes itself is fairly ridiculous. The user group on Google died about a year ago and the two main scientists who developed the software seem to either be reworking it or have moved on to other things. Some of the help suggestions involve one of the builders of the software tossing off stuff like "compiling RevBayes from the source code resolves this." I have then gone off on the really weird tangent of trying to compile RevBayes from the source code, only to find that the process depends on downloading a Linux application called cygwin that runs on command line from terminal, a whole area where I really just suck. So, the only way to resolve a huge bug in a software platform that I need to use is to learn an entire area of interaction with technology in which I have zero training. For people fluent in all of these bizarre-seeming routines, it's the easiest thing in the world. But for me, it's like being dropped in Athens in 700 BCE and having to learn Euclid.
I am currently running a fairly simple Bayesian divergence time estimate for my taxon, after finally getting it to actually run at all, and the ETA, running on my work issued MacBook Pro, is 300 hrs from now. And it has already been running the burn-in tuning of the tree proposals for six days. Meanwhile, I have also been troubleshooting a kind of useful phylogenetic tree plotting program also designed by the RevBayes team, that runs in R, called RevGadgets. The problem is that RevGadgets was written a long time ago and has not been updated, while its core dependency, ggtree, has been updated this month, and in turn uses a dependency, dplyr, which also has a recent update. It took me two days to unravel the simple fix of getting RevGadgets to run by uninstalling the new version of dplyr and installing the most recent version before that.
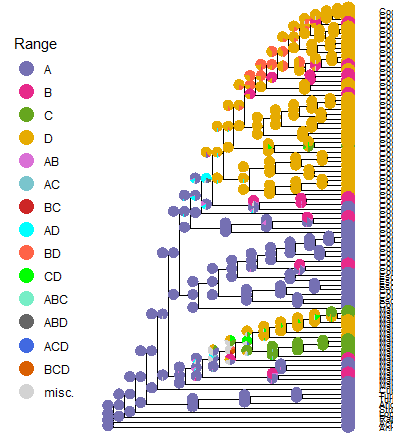 The figure above is a (very) rough draft of what I have been trying to produce. Taxon names are cut off to protect the innocent. haha. The figure shows the most likely ancestral geographical ranges of my taxa, with color coded pies on the tree nodes, showing the top three most likely ancestral ranges (that's why the pie nodes are vertically stacks, before each node). The ranges are A, eastern Mexico; B, the Sonoran Desert; C, the Sinaloan Gulf Coast; D, Baja California. The other colors in the pies are combined ranges. The tree is a time calibrated phylogeny (time scale not added to the figure yet, but the oldest node is about 13 million years ago). The idea is to get to some inference around the geospatial evolutionary history of my taxa. The model was built using epochs based on the most likely geological history of the areas used. For example, the eastern Mexico range pre-dates the Sonoran Desert region by about 5 million years. Another important event: the rifting of the Baja peninsula from the mainland, which supposedly started about 5 million years ago.
This project has been all consuming, really, the past couple of weeks. I currently have three computers running in my apartment, and I'm running a RevBayes job remotely on Agave, that is scheduled for six days.
It's a good thing I took a break from Facebook for the entire month of May. It's been the only way to focus on this project as well as devoting time and attention to the relationship with M, let alone my teaching job. I checked in briefly on music birthdays for today, and it's Brian Eno's 73rd birthday. It's also the 68th anniversary of Jazz at Massey Hall, the last time be bop pioneers Dizzy Gillespie, Charlie Parker, Bud Powell, Max Roach, and Charles Mingus played as a quintet.
I am taking today off, away from the constant buzz of the MacBook, ancient Toshiba (seven years old), sleek new ROG Zephyrus, and the distant, super genius Agave. I have to get outside. Probably going up to Angeles National Forest. Not sure. Time to get some distance from solving the mysteries of ancestral biogeography.
The figure above is a (very) rough draft of what I have been trying to produce. Taxon names are cut off to protect the innocent. haha. The figure shows the most likely ancestral geographical ranges of my taxa, with color coded pies on the tree nodes, showing the top three most likely ancestral ranges (that's why the pie nodes are vertically stacks, before each node). The ranges are A, eastern Mexico; B, the Sonoran Desert; C, the Sinaloan Gulf Coast; D, Baja California. The other colors in the pies are combined ranges. The tree is a time calibrated phylogeny (time scale not added to the figure yet, but the oldest node is about 13 million years ago). The idea is to get to some inference around the geospatial evolutionary history of my taxa. The model was built using epochs based on the most likely geological history of the areas used. For example, the eastern Mexico range pre-dates the Sonoran Desert region by about 5 million years. Another important event: the rifting of the Baja peninsula from the mainland, which supposedly started about 5 million years ago.
This project has been all consuming, really, the past couple of weeks. I currently have three computers running in my apartment, and I'm running a RevBayes job remotely on Agave, that is scheduled for six days.
It's a good thing I took a break from Facebook for the entire month of May. It's been the only way to focus on this project as well as devoting time and attention to the relationship with M, let alone my teaching job. I checked in briefly on music birthdays for today, and it's Brian Eno's 73rd birthday. It's also the 68th anniversary of Jazz at Massey Hall, the last time be bop pioneers Dizzy Gillespie, Charlie Parker, Bud Powell, Max Roach, and Charles Mingus played as a quintet.
I am taking today off, away from the constant buzz of the MacBook, ancient Toshiba (seven years old), sleek new ROG Zephyrus, and the distant, super genius Agave. I have to get outside. Probably going up to Angeles National Forest. Not sure. Time to get some distance from solving the mysteries of ancestral biogeography.


Love reading your update!